ANOVA
What ANOVA does
- ANalysis Of VAriance
- do multiple (more than 2) populations have the same mean?
- different types of variance are used to figure this out (sums of squares)
- null hypothesis: all groups do have the same mean
- alternative hypothesis: at least one mean differs from the others
- a low p-value means that the null is rejected, i.e. that at least one mean differs significantly
- functions:
aov();oneway.test; …
Assumptions
- data points must be independent
- all populations must be (approximately) normally distributed
- exploring normality: same as for T-test (with all groups)
- all groups must have similar variance (homoscedasticity)
Some data:
file.exists("anova_data.txt")## [1] TRUEdf = read.table("anova_data.txt", header = FALSE)
colnames(df)## [1] "V1" "V2" "V3" "V4"colnames(df) = c("A", "B", "C", "D")
head(df)## A B C D
## 1 0.35304697 4.983129 1.7420254 3.4038870
## 2 -0.92555558 4.479949 1.0426776 -1.1661505
## 3 1.47636942 4.054450 -1.8529604 0.5629787
## 4 -0.65262182 3.926977 0.5773551 1.7024414
## 5 0.87934474 5.626965 1.2850280 3.6237300
## 6 0.02417173 2.180872 1.2049257 0.6112000A boxplot (or violin plot) can help to get a first impression. Do the groups overlap?
boxplot(df, col="green", main="Comparison of four datasets")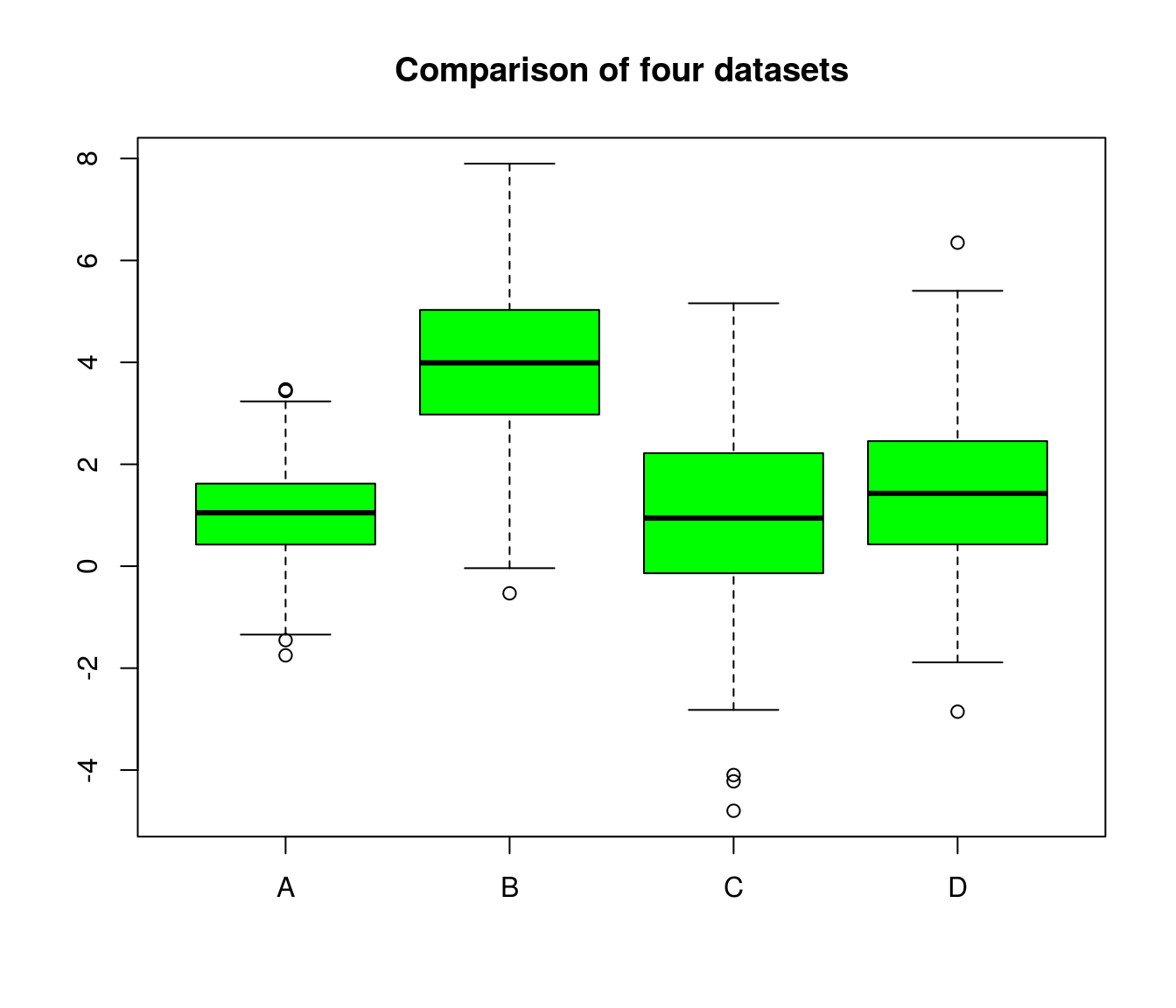
summary(df)## A B C D
## Min. :-1.7489 Min. :-0.5317 Min. :-4.7983 Min. :-2.8548
## 1st Qu.: 0.4257 1st Qu.: 2.9825 1st Qu.:-0.1313 1st Qu.: 0.4384
## Median : 1.0482 Median : 3.9907 Median : 0.9446 Median : 1.4285
## Mean : 1.0204 Mean : 3.9939 Mean : 0.9602 Mean : 1.4527
## 3rd Qu.: 1.6174 3rd Qu.: 5.0288 3rd Qu.: 2.2157 3rd Qu.: 2.4535
## Max. : 3.4694 Max. : 7.8987 Max. : 5.1594 Max. : 6.3481suppressMessages(library(vioplot)) # load library silently
vioplot(df$A, df$B, df$C, df$D, col = "gold", names = c("g1", "g2", "g3", "g4"))
title("Comparison of four groups", font.main = 1) 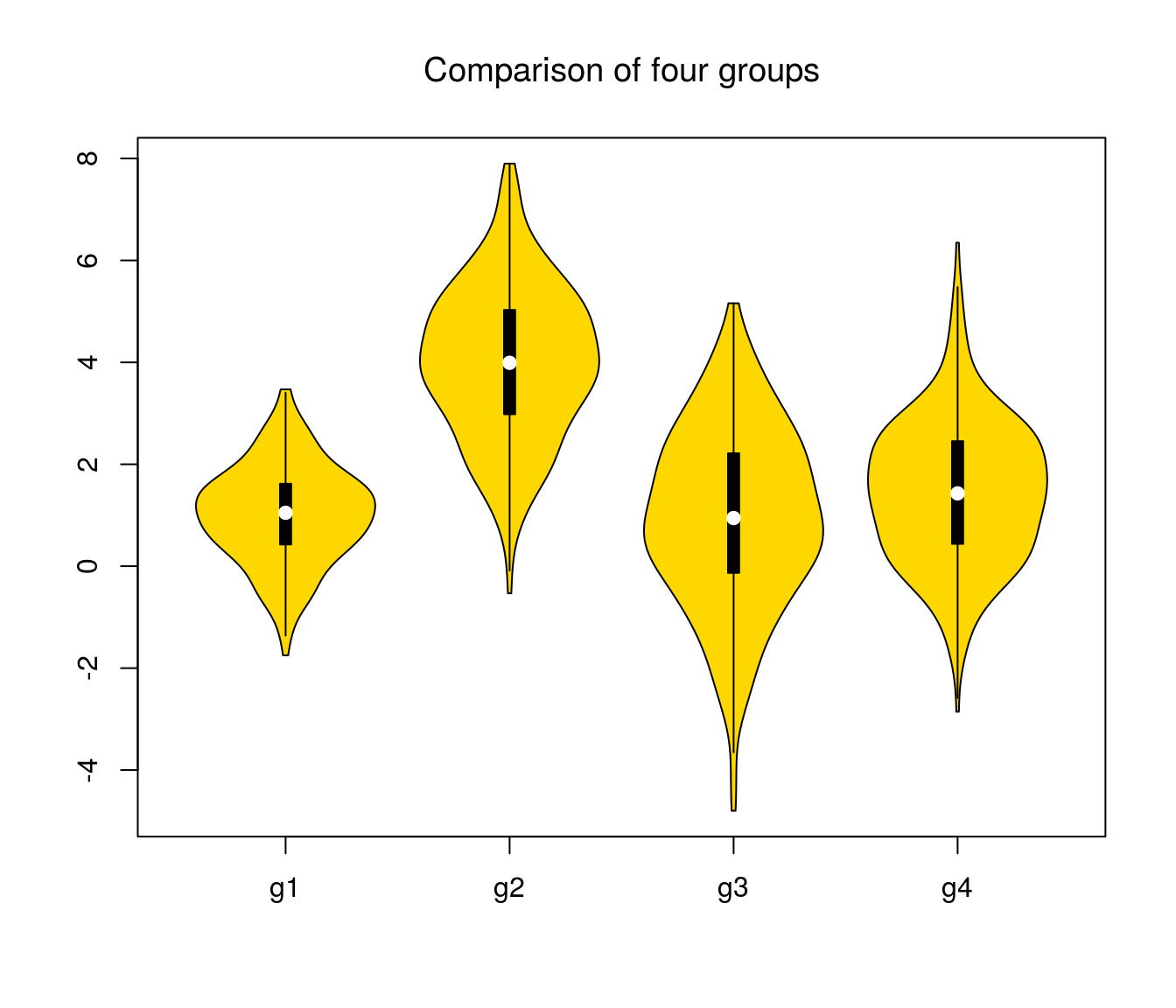
Checking for homoscledasticity
- Look at the boxplot!
- use
bartlett.test(but might be significant for large datasets, even if the differences are small)
Running the ANOVA using aov()
Some functions (e.g. aov() need stacked data as input:
stacked <- stack(df)
colnames(stacked) <- c("values", "groups")
head(stacked)## values groups
## 1 0.35304697 A
## 2 -0.92555558 A
## 3 1.47636942 A
## 4 -0.65262182 A
## 5 0.87934474 A
## 6 0.02417173 Atail(stacked)## values groups
## 1195 3.5114304 D
## 1196 1.1461898 D
## 1197 -0.5427561 D
## 1198 2.3259909 D
## 1199 1.4033644 D
## 1200 0.5924236 Dstr(stacked)## 'data.frame': 1200 obs. of 2 variables:
## $ values: num 0.353 -0.926 1.476 -0.653 0.879 ...
## $ groups: Factor w/ 4 levels "A","B","C","D": 1 1 1 1 1 1 1 1 1 1 ...Running aov():
- Low p-values: \(H_0\) to be rejected –> at least one mean differs significantly
res <- aov(values ~ groups, data = stacked)
summary(res) # reject the null## Df Sum Sq Mean Sq F value Pr(>F)
## groups 3 1870 623.4 303 <2e-16 ***
## Residuals 1196 2461 2.1
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Which group differs?
- run post-hoc test
- Tukey’s HSD (Honest Significant Difference)
- the Tukey test automatically corrects for multiple testing in a smart way
tuk <- TukeyHSD(res)
tuk## Tukey multiple comparisons of means
## 95% family-wise confidence level
##
## Fit: aov(formula = values ~ groups, data = stacked)
##
## $groups
## diff lwr upr p adj
## B-A 2.97345793 2.6721526 3.2747632 0.0000000
## C-A -0.06021073 -0.3615160 0.2410946 0.9557379
## D-A 0.43228489 0.1309796 0.7335902 0.0013327
## C-B -3.03366865 -3.3349740 -2.7323633 0.0000000
## D-B -2.54117303 -2.8424783 -2.2398677 0.0000000
## D-C 0.49249562 0.1911903 0.7938009 0.0001644plot(tuk)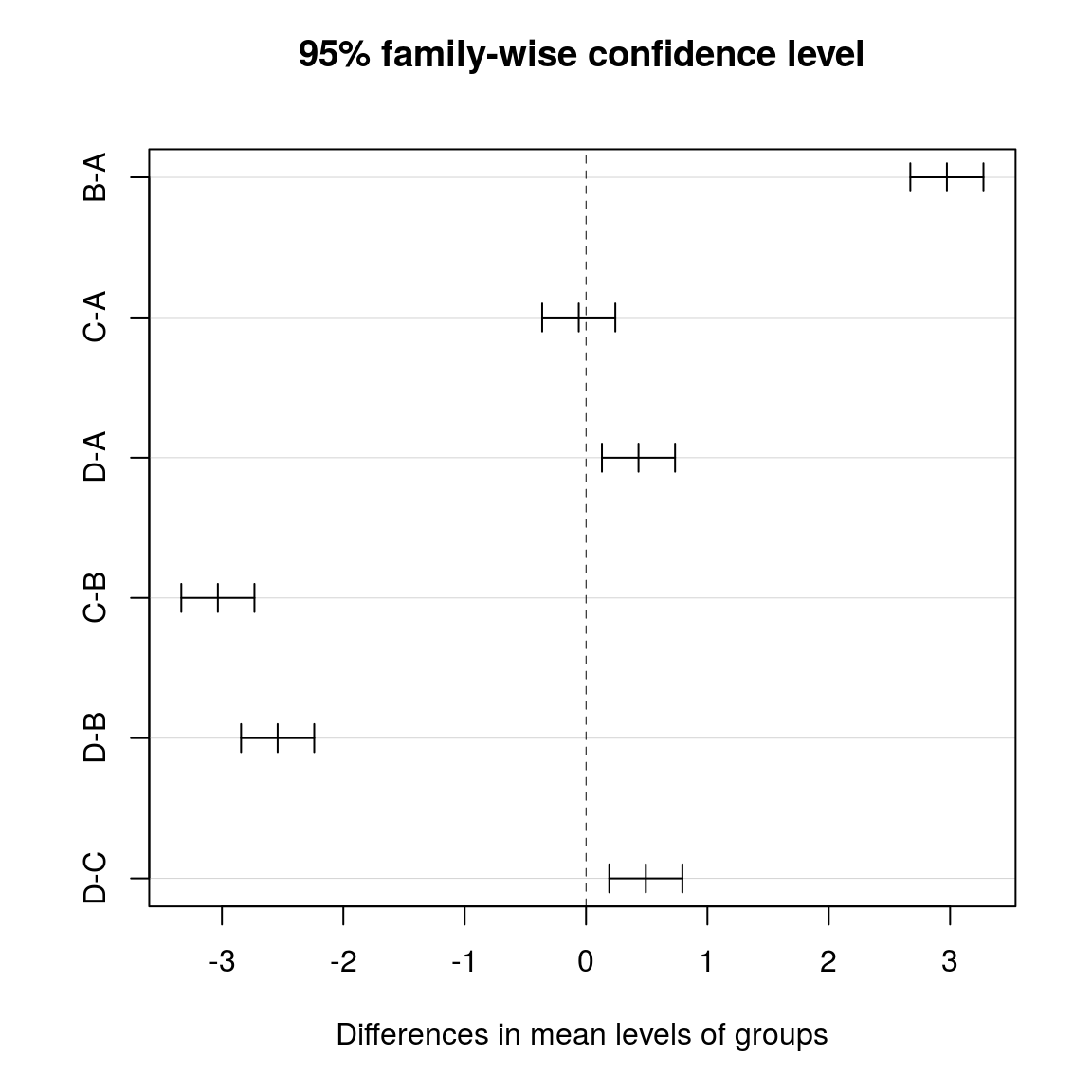
Running the ANOVA using oneway.test()
Which one to choose?
oneway.test()can correct for nonhomogeneity of the variances (Welch correction)- no post hoc test available for
oneway.test()
data(InsectSprays) # get some R dataset
head(InsectSprays)## count spray
## 1 10 A
## 2 7 A
## 3 20 A
## 4 14 A
## 5 14 A
## 6 12 Astr(InsectSprays)## 'data.frame': 72 obs. of 2 variables:
## $ count: num 10 7 20 14 14 12 10 23 17 20 ...
## $ spray: Factor w/ 6 levels "A","B","C","D",..: 1 1 1 1 1 1 1 1 1 1 ...levels(InsectSprays$spray) # 6 levels ## [1] "A" "B" "C" "D" "E" "F"boxplot(count ~ spray, data = InsectSprays) # not normally distr. (assymmetric), different variance in the groups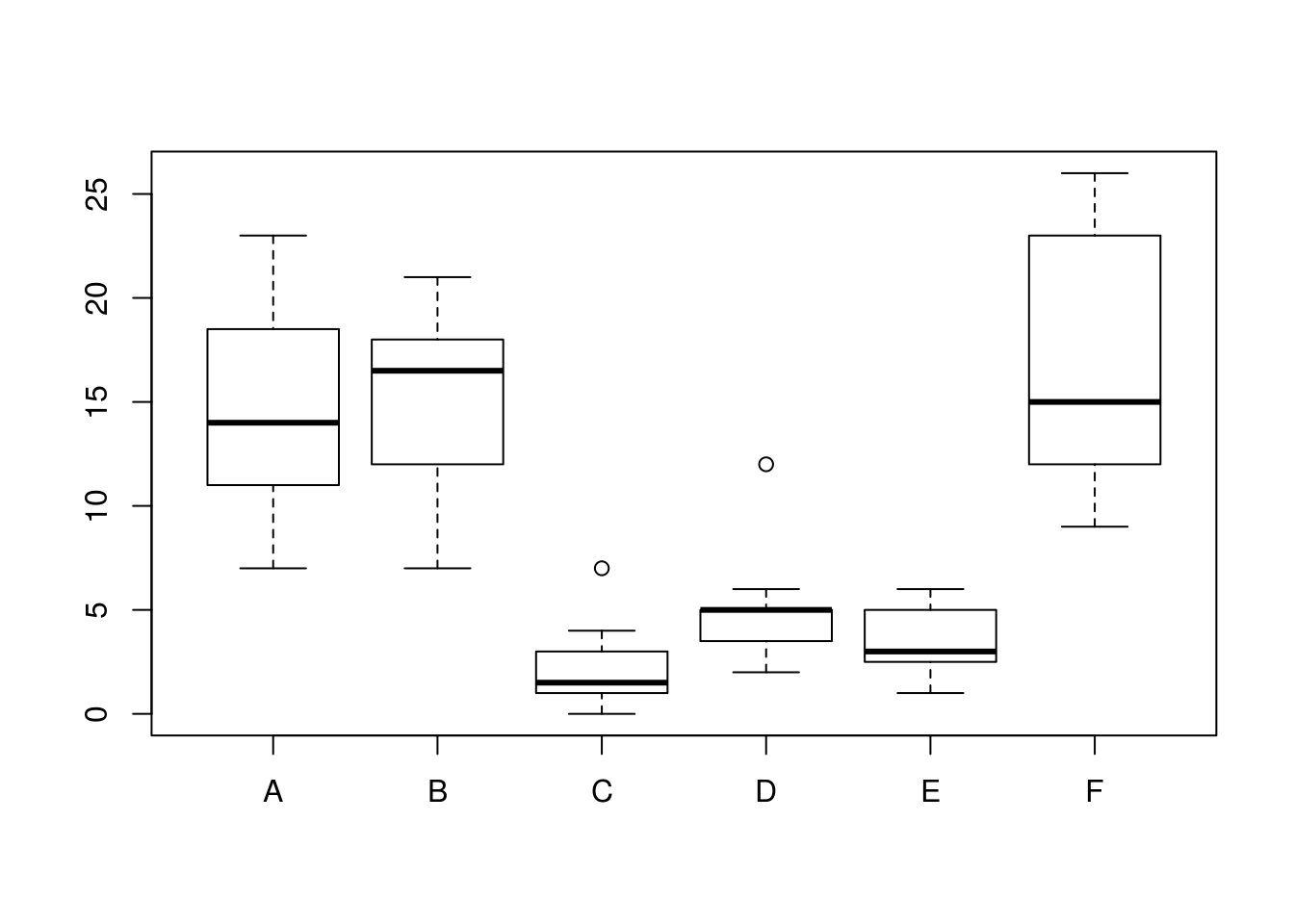
bartlett.test(count ~ spray, data = InsectSprays) # check for heteroscedasticity ##
## Bartlett test of homogeneity of variances
##
## data: count by spray
## Bartlett's K-squared = 25.96, df = 5, p-value = 9.085e-05oneway.test(count ~ spray, data = InsectSprays) # var.equal = FALSE is default!##
## One-way analysis of means (not assuming equal variances)
##
## data: count and spray
## F = 36.065, num df = 5.000, denom df = 30.043, p-value = 7.999e-12When the assumptions for ANOVA are not met, part I
Try data tranformation? (log, square root, …)
The same as for T-test
When the assumptions for ANOVA are not met, part II
Conduct a non-parametric test (rank sum test)
- Kruskal-Wallis test
- rank-based nonparametric test
- if the data is normally distributed, the ANOVA is better (has more power)
kruskal.test(df) # on data frame##
## Kruskal-Wallis rank sum test
##
## data: df
## Kruskal-Wallis chi-squared = 480.82, df = 3, p-value < 2.2e-16kruskal.test(values ~ groups, data = stacked) # on stacked data##
## Kruskal-Wallis rank sum test
##
## data: values by groups
## Kruskal-Wallis chi-squared = 480.82, df = 3, p-value < 2.2e-16