Multidimensional Scaling plots
What does MDS do?
- roughly spoken, it shows a 2D (3D) projection of a multidimensional cloud in such a way that the relative distances between all points are preserved
MDS plots using cmdscale
Load some data:
expr = get(load("RPKM_means_DOG_DMSO.RData"))
dim(expr)## [1] 8 24607rownames(expr)## [1] "1d_mean_DMSO" "5d_mean_DMSO" "10d_mean_DMSO" "20d_mean_DMSO"
## [5] "1d_mean_DOG" "5d_mean_DOG" "10d_mean_DOG" "20d_mean_DOG"We have to tell cmdscale what distance measure we want to use.
The function cmdscale takes an object of class dist as input:
distance = dist(expr, method = "euclidean") # dist computes distances between the rows of a matrix.
class(distance) ## [1] "dist"length(distance)## [1] 28scaling <- cmdscale(distance, k = 2, eig = TRUE) # create 2D-plot
str(scaling)## List of 5
## $ points: num [1:8, 1:2] 80879 -14010 -30137 -58733 111974 ...
## ..- attr(*, "dimnames")=List of 2
## .. ..$ : chr [1:8] "1d_mean_DMSO" "5d_mean_DMSO" "10d_mean_DMSO" "20d_mean_DMSO" ...
## .. ..$ : NULL
## $ eig : num [1:8] 2.89e+10 7.54e+09 2.50e+09 1.24e+09 4.22e+08 ...
## $ x : NULL
## $ ac : num 0
## $ GOF : num [1:2] 0.89 0.89x <- scaling$points[,1]
y <- scaling$points[,2]
plot(x,y)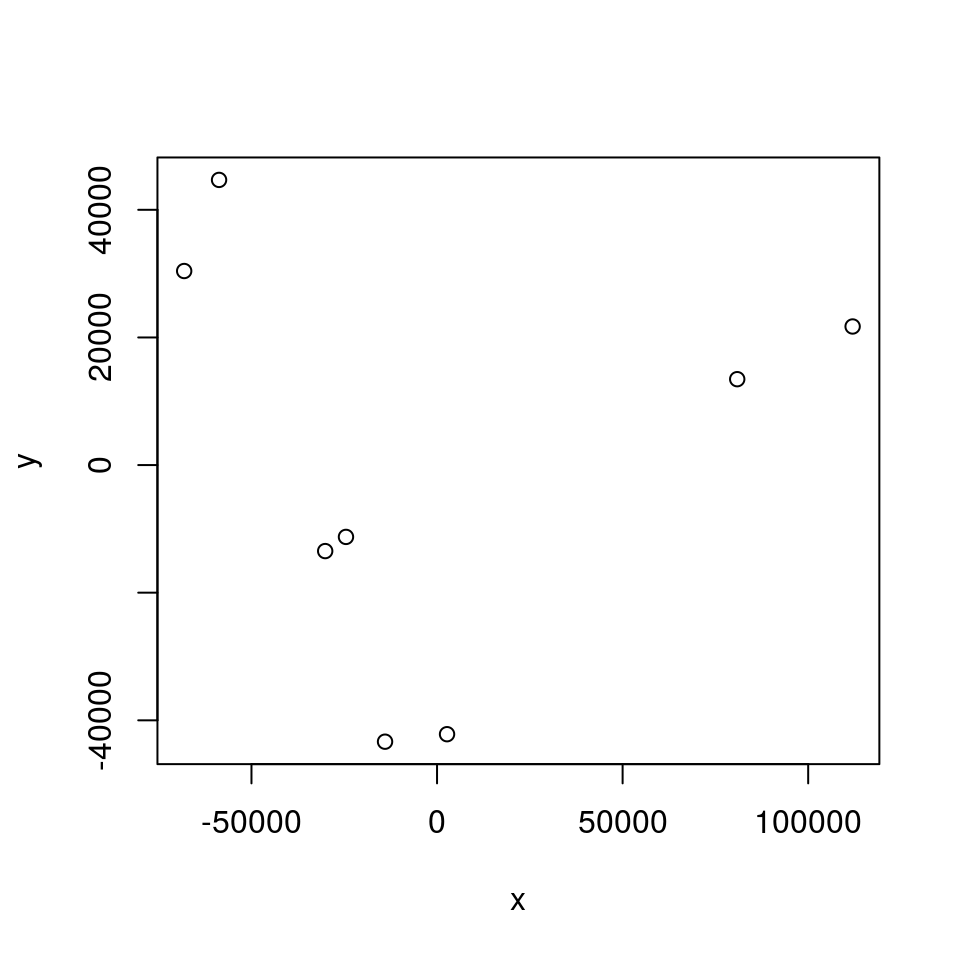
Improved plot
Colouring the points according to time (1d, 5d, 10d, 20d) might be a good idea:
rownames(expr)## [1] "1d_mean_DMSO" "5d_mean_DMSO" "10d_mean_DMSO" "20d_mean_DMSO"
## [5] "1d_mean_DOG" "5d_mean_DOG" "10d_mean_DOG" "20d_mean_DOG"colorvec = rep("black", nrow(expr))
grep("1d", rownames(expr)) ## [1] 1 5colorvec[grep("1d", rownames(expr))] = "blue"
colorvec[grep("5d", rownames(expr))] = "red"
colorvec[grep("10d", rownames(expr))] = "green"
colorvec[grep("20d", rownames(expr))] = "brown"
colorvec## [1] "blue" "red" "green" "brown" "blue" "red" "green" "brown"cbind(rownames(expr), colorvec)## colorvec
## [1,] "1d_mean_DMSO" "blue"
## [2,] "5d_mean_DMSO" "red"
## [3,] "10d_mean_DMSO" "green"
## [4,] "20d_mean_DMSO" "brown"
## [5,] "1d_mean_DOG" "blue"
## [6,] "5d_mean_DOG" "red"
## [7,] "10d_mean_DOG" "green"
## [8,] "20d_mean_DOG" "brown"Shaping the points according to medium (look here for possible shapes:
rownames(expr) ## [1] "1d_mean_DMSO" "5d_mean_DMSO" "10d_mean_DMSO" "20d_mean_DMSO"
## [5] "1d_mean_DOG" "5d_mean_DOG" "10d_mean_DOG" "20d_mean_DOG"pchvec = rep(1, nrow(expr))
grep("DMSO", rownames(expr))## [1] 1 2 3 4pchvec[grep("DMSO", rownames(expr))] = 17
pchvec[grep("DOG", rownames(expr))] = 18
pchvec## [1] 17 17 17 17 18 18 18 18cbind(rownames(expr), pchvec)## pchvec
## [1,] "1d_mean_DMSO" "17"
## [2,] "5d_mean_DMSO" "17"
## [3,] "10d_mean_DMSO" "17"
## [4,] "20d_mean_DMSO" "17"
## [5,] "1d_mean_DOG" "18"
## [6,] "5d_mean_DOG" "18"
## [7,] "10d_mean_DOG" "18"
## [8,] "20d_mean_DOG" "18"Plot it:
plot(x, y, col = colorvec, pch = pchvec, xlab = "", ylab = "")
legend("bottomright", c("1d", "5d", "10d", "20d"), col = c("blue", "red", "green", "brown"), pch = 16)
legend("top", c("DMSO", "DOG"), pch = c(17, 18), col = "black")
title("MDS plot, time & medium")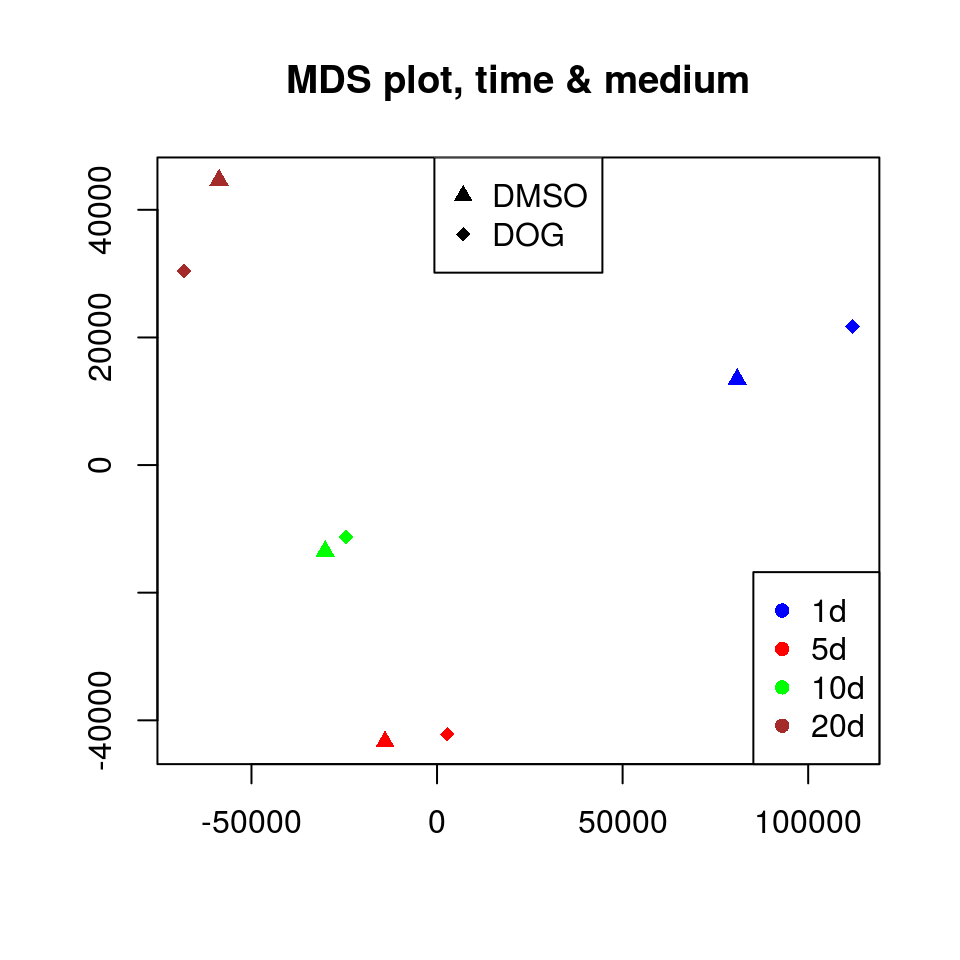
What about showing the distance between the genes?:
Turn the matrix around (“transpose”) in the dist command:
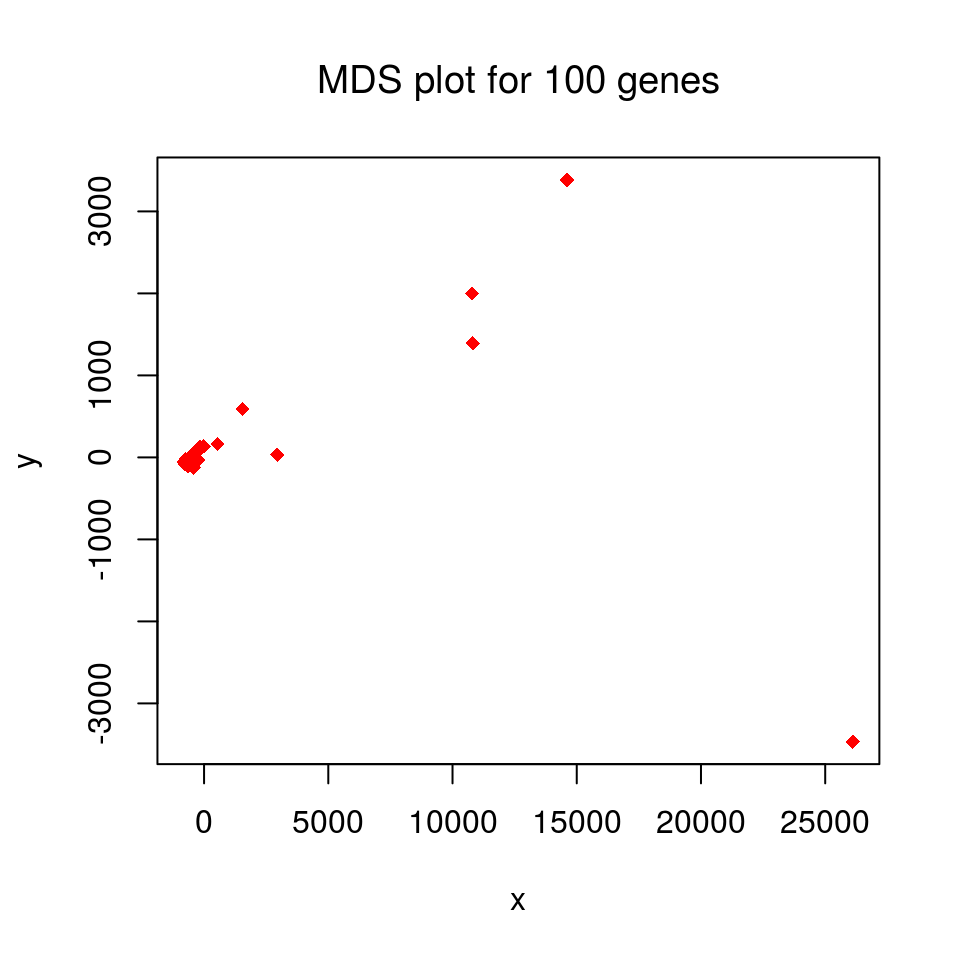
n = 100 # include that many genes
sample(1:ncol(expr), n) # choose randomly ## [1] 6797 5328 7913 3422 19903 10974 19893 8962 816 17612 14198
## [12] 4242 10144 21478 3141 5955 6373 1901 13716 20860 5890 16108
## [23] 7098 10881 10889 22785 8445 21149 10412 8309 7262 12146 1588
## [34] 19917 17043 10926 14193 9824 18518 23976 7387 20689 11162 12175
## [45] 12940 6491 15908 19550 17763 23515 17777 1230 9981 3445 340
## [56] 24026 7864 4788 606 13970 9048 9345 7137 19995 20804 13158
## [67] 12300 11324 18966 322 12107 9458 14960 2871 5307 15033 14567
## [78] 8741 21710 14682 19659 19619 16219 15428 11972 12961 8173 1451
## [89] 23046 12211 2138 17220 4743 4227 4514 4739 10961 18347 5250
## [100] 9830set.seed(76)
small = expr[, sample(1:ncol(expr), n)] # select n genes randomly
distance = dist(t(small), method = "euclidean") # dist computes distances between the rows of a matrix.
length(distance)## [1] 4950scaling <- cmdscale(distance, k = 2, eig = TRUE) # create 2D-plot
str(scaling)## List of 5
## $ points: num [1:100, 1:2] -784 -739 -800 -437 -800 ...
## ..- attr(*, "dimnames")=List of 2
## .. ..$ : chr [1:100] "R173.1" "T28H10.3" "F49E12.7" "C17E4.3" ...
## .. ..$ : NULL
## $ eig : num [1:100] 1.19e+09 3.01e+07 2.63e+06 6.86e+05 2.50e+05 ...
## $ x : NULL
## $ ac : num 0
## $ GOF : num [1:2] 0.997 0.997x <- scaling$points[,1]
y <- scaling$points[,2]
maintxt = paste("MDS plot for", n, "genes")
plot(x, y, col = "red", pch = 18, main = maintxt, font.main = 1)