What can `seqinr’ do?
- Exploratory data analysis and data visualization for biological sequence (DNA and protein) data
- R documentation chapter
DNA
library(seqinr)
dna = read.fasta("seqDNA.fasta", seqtype = "DNA", as.string = TRUE, forceDNAtolower = FALSE)
class(dna)
## [1] "list"
length(dna)
## [1] 3
names(dna)
## [1] "AK002358.PE1" "HSU78678.PE1" "RNU73525.PE1"
dna2 = read.fasta("seqDNA.fasta")
GC(dna2[[1]])
## [1] 0.5508982
write.fasta(dna[[3]], names="dna3", file.out="dna3.fasta", open = "w", nbchar = 60)
file.exists("dna3.fasta")
## [1] TRUE
aln <- read.alignment("alignment.phy", format = "phylip", forceToLower = TRUE)
names(aln)
## [1] "nb" "nam" "seq" "com"
aln$nb
## [1] 39
aln$seq[[1]]
## [1] "----iafnmamevririlndcstygtnnfkvlapsnsstaaavtfitfvqdlkrktktwgpmielcasgektlerfryqfpddwlysdqlkgewsafneilkrkndsiqeqlaglqlkivaedkivenkindviqewettrpvrgdipasealsainvfdqrltrvqeeydlvcrakealdldlirhtrlepifeelrdlkavwtalsgiwsqvselrdlswatvqprklrqqldglltstkemptrmrqyaafeyvqerlkgllkantllselksdalkdrhwkqlfkvlrvsnpptlnlmtlgivydmdlkhnenlikevivqaqgemaleeyirqvketwtaytldlvnyqnkcrlirgwddlfnkcsenlnsltamrlspyykgiftgsadikhllpvesarfnnintefqavmkkvykspfvvdvmnipgiqkslerladllnkiqkalgeylererasfprfyfvgdedlleiignskeilrimkhlkkmfagistiklddeltqilgmasregeevtfkdpimlkdypkindwltkleaemrnslalllcdavaelqafygtgeqleqkqfiewiekypaqlvtlaiqvawtasiedalqqqpptlnrpletirqgldlladvvltelaavtrhkcehlitelvhqrdvtrtliqqgvsdsrafswlyqmrfyldttvespldrlsirvadasfpygweylgvpdrlvqtpltdrcyltltqaldnqlggspfgpagtgkcktesvkalgvqlgrfvlvfccdetfdfqamgrifvglcqvgawgcfdefnrleerilsavsqqvqsiqqglatlvknpsaeidlvgksvkinknmgifittnpnyagrstlppnltklfrpmamtrpdreliaqvmlfsqgfrtaeslaskivpffnlcdeqlspqphydfglralkavlasagilkrerlqaassgdsdvdvvglsdstseqviliqsvtetivpklvaddvplltrfvnnmrvsfqtldvsfslladvfpgtdyipvnldnlreqidkvcterrlvqndrwiskilqlyqiqkiqhglmmvgpsgagktnawqvllaalerldgiegvsyvidpkamhkdalygtldpttrewtdglfthtlrkivddvrgesgkrhwivfdgdvdpewvenlnsvlddnklltlpngerlnlppnvrimfevehlkyatlatvsrcgmiwfsedvvdasmvcrhyldtlssvpldaeeddsreilgrrsdslipddsssailatqkqvsailepffaddgliasalthaesiehimdftvtralntlfslvnktarniieynmqhtdfpltperieqyvskrllvniiwafsgdakldlraemgdflrkqtgidlppliqgsslldydvavntgewsawqsrvptieieahsitasdvvvptidtirheevlyswlsehkplmlcgppgsgktmtlfsalrklpdmevvglnfssattpelllktfeqycefrktpngvilapvqlgrwlvvfcdeinlpatdkygtqrvisfirqlvecngywrtsdmawvkleriqfvgacnpptdpgrvplshrflrhaplimvdypgevslkqiygaynhamlkvipslraysgpltdamvslylasqkrfttdiqahyvyspreltrwvrgmyeairpleslsveglvrvwahealrlfqdrlvtedekqwtdkqidasamehfptinteealgrpilfsnwtsknyvpvdretlrehtkarlrvfyeeeldvplvlfndvldhvlridrvfrqvqghllligvsgsgkttlsrfvawmnglsifqikvsnkytgedfdedlrtvlrragckgekicfimdesnvldsgflermntllanaevpglfegdehsalmtackegsqrdglmldsheelyrwftqqvaknlhvvftmnppenglasraatspalfnrcvldwfgdwsdqalyqvgieftqtldldvpsydapdhfpiayrelemppihrtavvnalvhvhqslhqinqrlsrrqgsinhyvrlynekrdeleeqqrhlhvgldklrdtvtqveelrkslaikrtqlqakdaeaneklkrmvadqqeaeqkkaasieiqaalveqdrnieqrrsvvmadladaepavmdaqaavsnikrqhlqevrtmanppeavklamesvctllghkfdtwrnvqgiirrddfissivgfnterltkhvrdamkrdflsrpsfnyetvqraskacgplvkwviaqvryseildkveplrnevrsleeqaeqtkeqakmiikmiseleasierykeeyaaliretqaiktetervqskvdrsmklleslsseksrwelgsrtfdtemstivgdvllsagflayagffdqqyremmwqewsthlsdagikfkaelslpeylstaddrlswqsrslpsdnlctenaimikrfsryplvidptgqatnfllneykdrkitvtsfldeaflkvlesalrfgntlliqdvehldpilnpvlnkeirrtggrvlirlgnqdidfspaftmflstrdpsvefspdicsrvtfvnftmtrsslqsqsldqvlkverpdterkrtdlmkmqgefrlrlrtleklllqalnestgnildddkvidtletlkreaaeitrkveetdivmkeveevtaeylplaqacsavffileqlnlvnhfyqfslrffldifdhvlhhnpnlknvadhsgrrdillndlfltvyrrtsrallhrdhlmlavlltqvklrgieeigdeleflleagdgappsgavetrnsllsegqaqrlekfaqhplfkpvlnhiqqnedlwvpflqsatpetdvpypwepstrrlipytddrrltivlpa---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------"
Amino Acids
amino = read.fasta("seqAA.fasta", seqtype = "AA")
amino
## $A06852
## [1] "M" "P" "R" "L" "F" "S" "Y" "L" "L" "G" "V" "W" "L" "L" "L" "S" "Q"
## [18] "L" "P" "R" "E" "I" "P" "G" "Q" "S" "T" "N" "D" "F" "I" "K" "A" "C"
## [35] "G" "R" "E" "L" "V" "R" "L" "W" "V" "E" "I" "C" "G" "S" "V" "S" "W"
## [52] "G" "R" "T" "A" "L" "S" "L" "E" "E" "P" "Q" "L" "E" "T" "G" "P" "P"
## [69] "A" "E" "T" "M" "P" "S" "S" "I" "T" "K" "D" "A" "E" "I" "L" "K" "M"
## [86] "M" "L" "E" "F" "V" "P" "N" "L" "P" "Q" "E" "L" "K" "A" "T" "L" "S"
## [103] "E" "R" "Q" "P" "S" "L" "R" "E" "L" "Q" "Q" "S" "A" "S" "K" "D" "S"
## [120] "N" "L" "N" "F" "E" "E" "F" "K" "K" "I" "I" "L" "N" "R" "Q" "N" "E"
## [137] "A" "E" "D" "K" "S" "L" "L" "E" "L" "K" "N" "L" "G" "L" "D" "K" "H"
## [154] "S" "R" "K" "K" "R" "L" "F" "R" "M" "T" "L" "S" "E" "K" "C" "C" "Q"
## [171] "V" "G" "C" "I" "R" "K" "D" "I" "A" "R" "L" "C" "*"
## attr(,"name")
## [1] "A06852"
## attr(,"Annot")
## [1] ">A06852 183 residues"
## attr(,"class")
## [1] "SeqFastaAA"
# methods(class = "SeqFastaAA")
AAstat(amino[[1]])
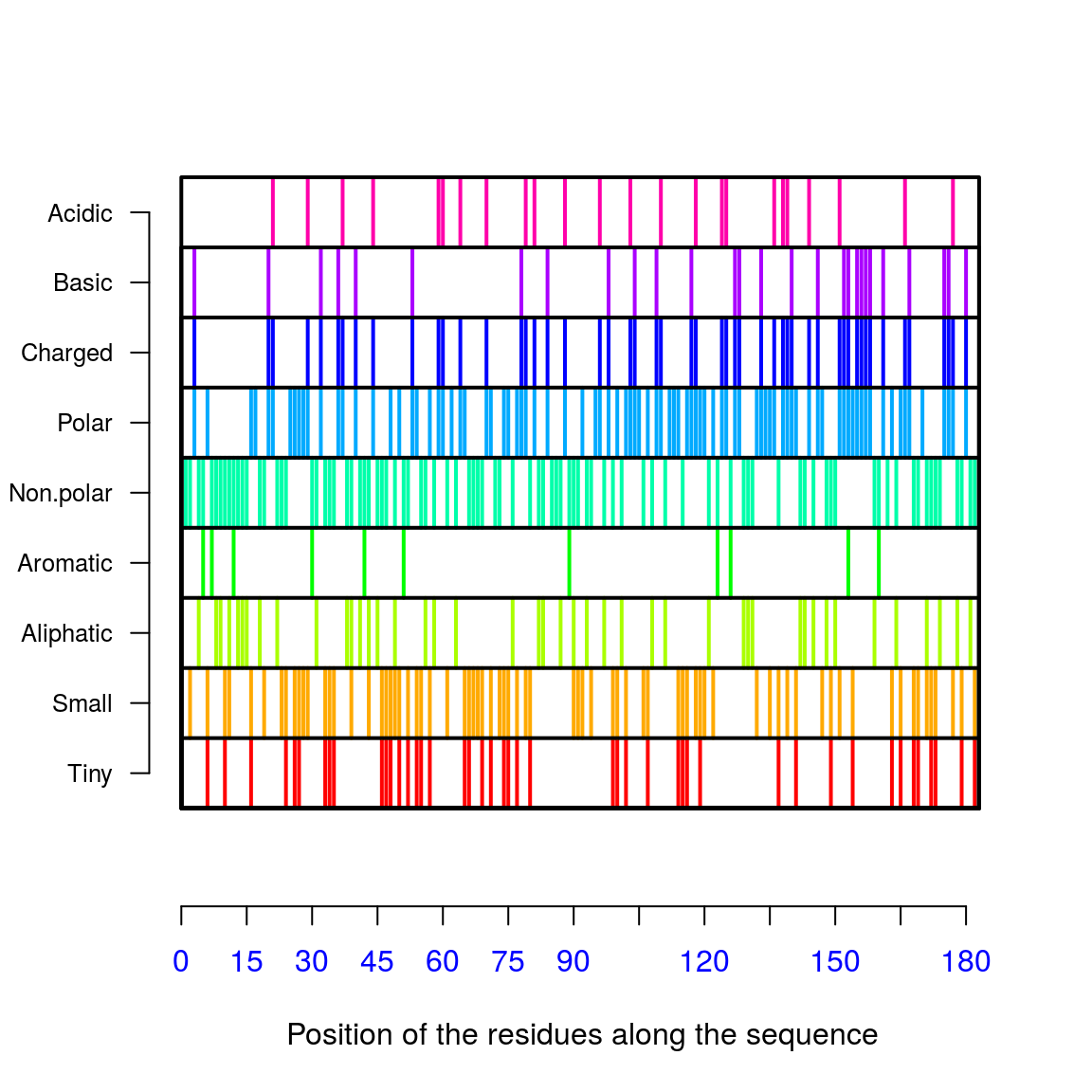
## $Compo
##
## * A C D E F G H I K L M N P Q R S T V W Y
## 1 8 6 6 18 6 8 1 9 14 29 5 7 10 9 13 16 7 6 3 1
##
## $Prop
## $Prop$Tiny
## [1] 0.2459016
##
## $Prop$Small
## [1] 0.4043716
##
## $Prop$Aliphatic
## [1] 0.2404372
##
## $Prop$Aromatic
## [1] 0.06010929
##
## $Prop$Non.polar
## [1] 0.4972678
##
## $Prop$Polar
## [1] 0.4972678
##
## $Prop$Charged
## [1] 0.284153
##
## $Prop$Basic
## [1] 0.1530055
##
## $Prop$Acidic
## [1] 0.1311475
##
##
## $Pi
## [1] 8.534902
names(amino)
## [1] "A06852"
summary(amino)
## Length Class Mode
## A06852 183 SeqFastaAA character
getSequence(amino)
## [[1]]
## [1] "M" "P" "R" "L" "F" "S" "Y" "L" "L" "G" "V" "W" "L" "L" "L" "S" "Q"
## [18] "L" "P" "R" "E" "I" "P" "G" "Q" "S" "T" "N" "D" "F" "I" "K" "A" "C"
## [35] "G" "R" "E" "L" "V" "R" "L" "W" "V" "E" "I" "C" "G" "S" "V" "S" "W"
## [52] "G" "R" "T" "A" "L" "S" "L" "E" "E" "P" "Q" "L" "E" "T" "G" "P" "P"
## [69] "A" "E" "T" "M" "P" "S" "S" "I" "T" "K" "D" "A" "E" "I" "L" "K" "M"
## [86] "M" "L" "E" "F" "V" "P" "N" "L" "P" "Q" "E" "L" "K" "A" "T" "L" "S"
## [103] "E" "R" "Q" "P" "S" "L" "R" "E" "L" "Q" "Q" "S" "A" "S" "K" "D" "S"
## [120] "N" "L" "N" "F" "E" "E" "F" "K" "K" "I" "I" "L" "N" "R" "Q" "N" "E"
## [137] "A" "E" "D" "K" "S" "L" "L" "E" "L" "K" "N" "L" "G" "L" "D" "K" "H"
## [154] "S" "R" "K" "K" "R" "L" "F" "R" "M" "T" "L" "S" "E" "K" "C" "C" "Q"
## [171] "V" "G" "C" "I" "R" "K" "D" "I" "A" "R" "L" "C" "*"
seq = paste(getSequence(amino)[[1]], collapse="")
seq
## [1] "MPRLFSYLLGVWLLLSQLPREIPGQSTNDFIKACGRELVRLWVEICGSVSWGRTALSLEEPQLETGPPAETMPSSITKDAEILKMMLEFVPNLPQELKATLSERQPSLRELQQSASKDSNLNFEEFKKIILNRQNEAEDKSLLELKNLGLDKHSRKKRLFRMTLSEKCCQVGCIRKDIARLC*"
